|
[Oct. 2023]
Happy to welcome two new PhD students whom I will be co-supervising for the next three years: Siegfried Dubois will explore pangenome graphs to characterize structural variants and Nicolas Maurice will work on metagenome assembly of complex microbial communities associated to plants, taking advantage of latest high fidelity long read technologies.
|

|
[July 2023]
SVJedi-graph, our new Structural Variant genotyping tool is published in Bioinformatics ! Look at the paper here and meet Sandra and me at ISMB/ECCB 2023 in Lyon (France) for the corresponding talk.
|
|
[July 2023]
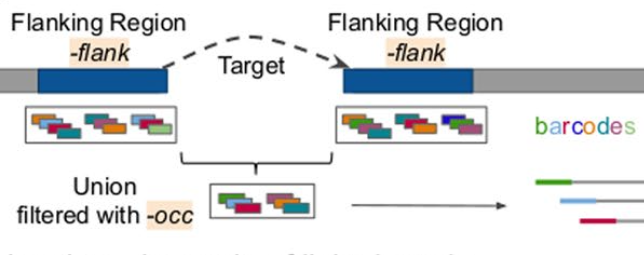
MTG-Link, our local assembly tool dedicated to linked-reads is finally published in BMC Bioinformatics ! Look at the github repository and read the paper which presents various biological applications, such as the reconstruction of the sequence of a locus of interest and of dark regions, gap-filling in draft assemblies, as well as alternative allele reconstruction of large Structural Variants.
|
|
[Sept 2022]
Release of a novel SV genotyping tool : SVJedi-graph improves over SVJedi to genotype Structural Variants with long reads thanks to the use of a variation graph to represent the SVs, and in particular close or overlapping SVs.
|

|
[May 2022]
As co-chair of the PC, I am very proud of this great program at JOBIM 2022 in Rennes (July 5-8) ! Don't delay in registering, the French bioinformatics community is eager to meet again in person ! (Update: JOBIM is full!)
|
|
[March 2022]
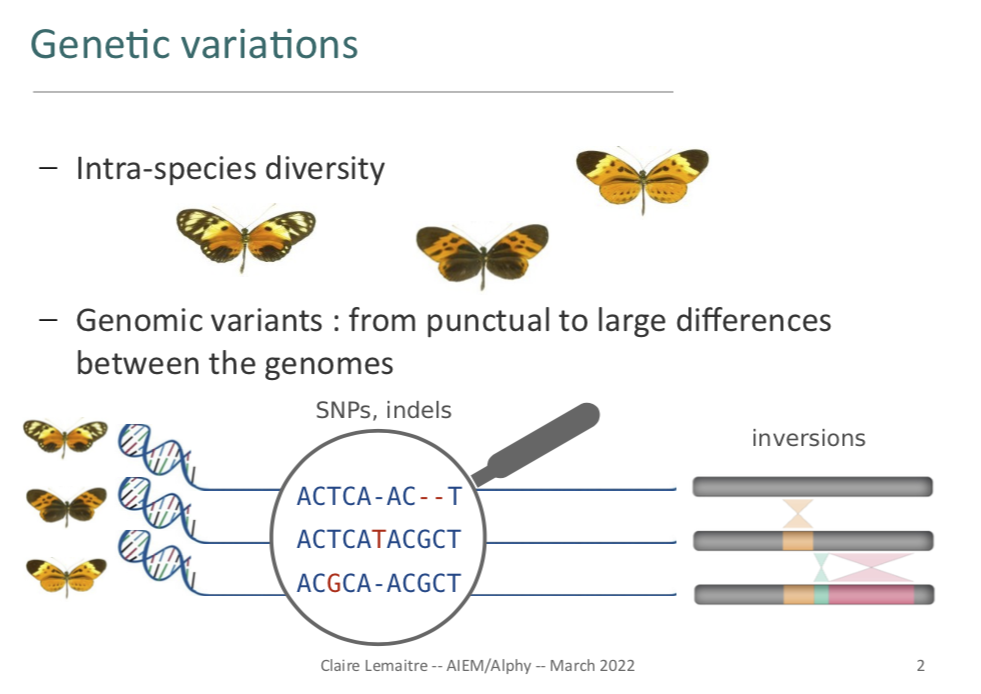
Very happy to give an invited talk at the annual meeting of the GDR AIEM and Alphy working group (GDR BIM) about methods for Structural Variation detection and analyses, with a focus on large insertion variants (see my slides)
|

|
[Dec 2021]
I just defended my HDR ("Habilitation à Diriger des recherches", or accreditation to supervise research, a final french diploma). The title of my manuscript is "Bioinformatics methods for studying Structural Variations with sequencing data" (look at my manuscript in french and the slides of my defense in english).
|

|
[Oct 2021]
Here we go for JOBIM 2022 in Rennes ! Happy to be co-chair of the Program Committee of this annual event that federates the French bioinformatics community.
|
|
[March 2021]
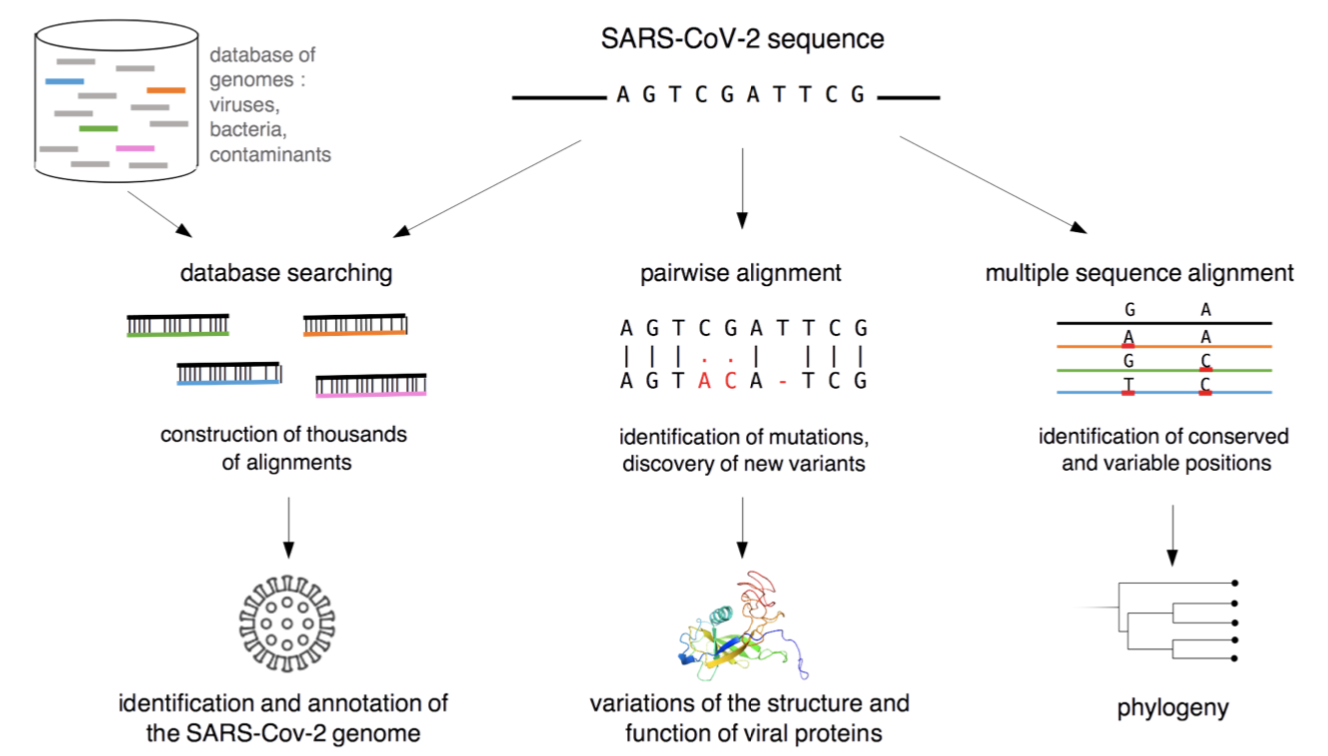
SARS-CoV-2 and bioinformatics: with the GDR BIM, we wrote a science popularization report on how Bioinformatics is playing a key role in the study of the virus and its origins.
|
|
[Dec 2020]
Two PhD defenses to come in December (4th and 11th), that I am proud to have supervised ! Structural Variations are in the spotlight with Lolita Lecompte and Wesley Delage !
|
|
[Nov 2020]
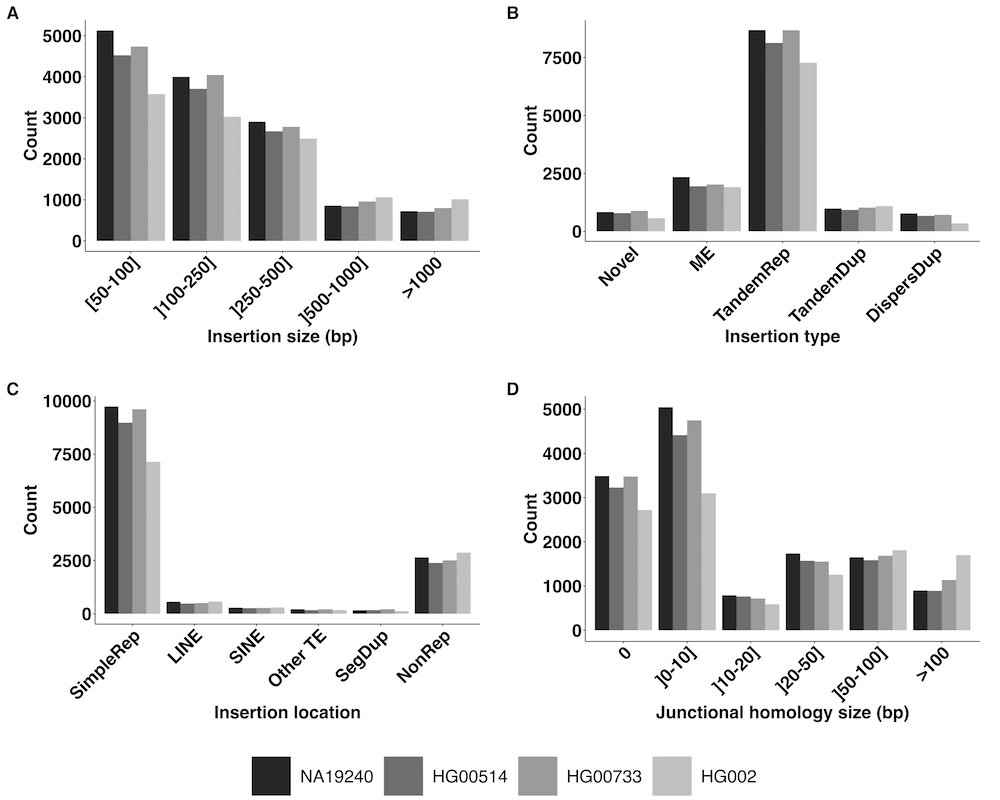
Towards a better understanding of the low recall of insertion
variants with short-read based variant callers. Our results on large human insertion variant characterization and how their features impact the recall of short-read based SV callers are out in BMC Genomics
|
|
[Sept 2020]
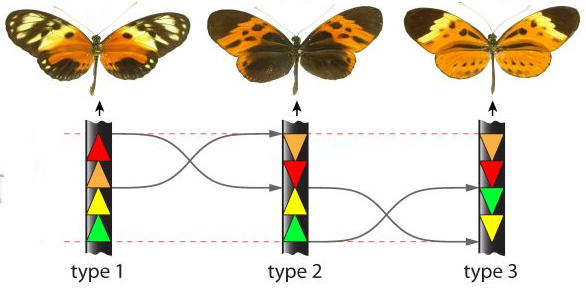
Welcoming Pierre Morisse for a 2-year postdoctoral fellowship in our team to develop novel methods for Structural Variation detection with linked-reads sequencing data, and applications to the mimetic butterfly Heliconius numata.
|

|
[July 2020]
Finding viral DRJ excision sites with sequencing data : a method I developped a long time ago to refine breakpoints is finally published within the analysis of two genomes of parasitic wasps having peculiar endogenous viruses. Paper in BMC Biology. The method, DrjBreakpointFinder, is available on github.
|
|
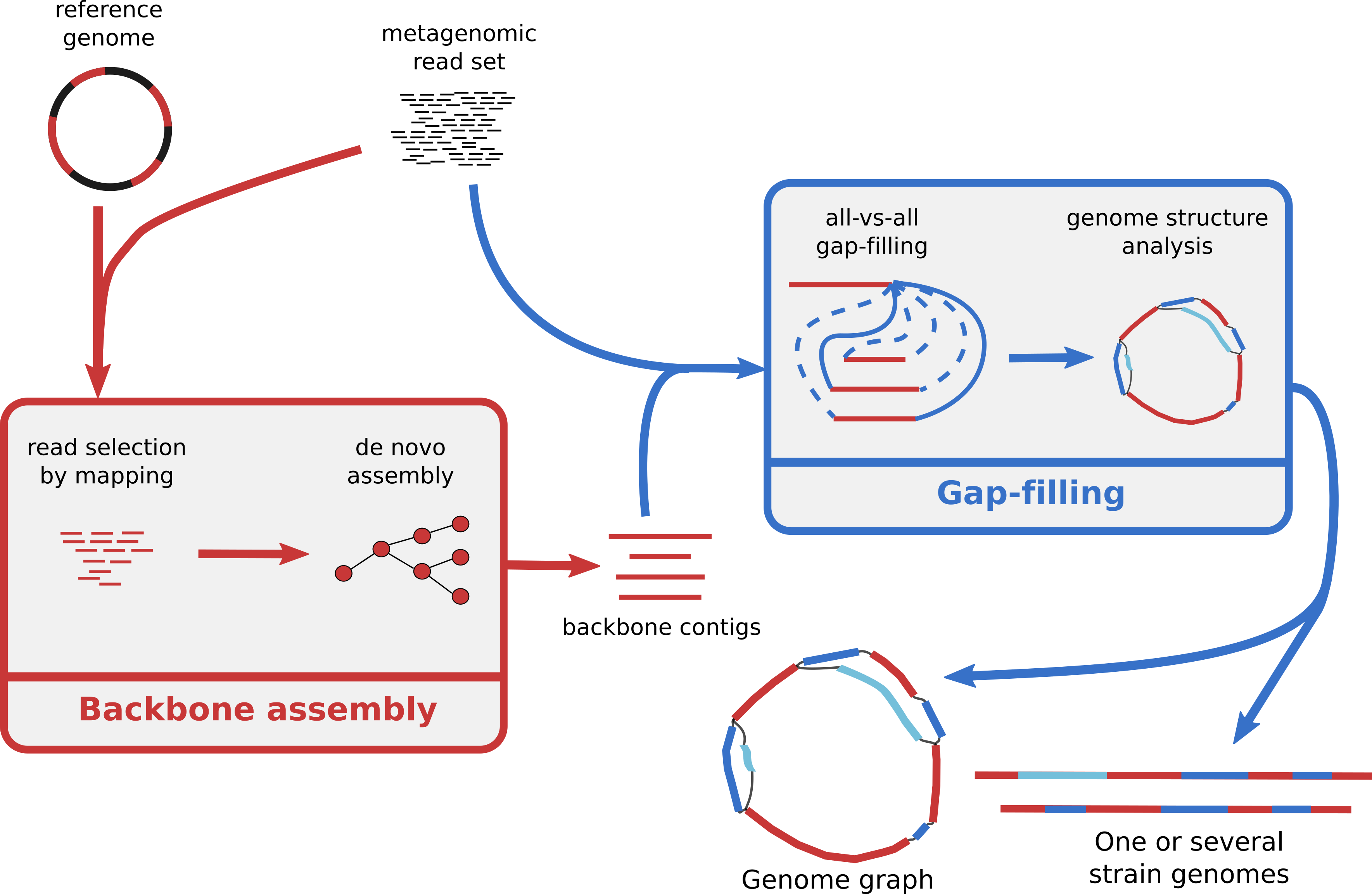
[July 2020]
MinYS is published in NAR Genomics and Bioinformatics ! : MinYS (for Mine Your Symbiont) is a new software developped with a former PhD student, Cervin Guyomar, for targeted genome assembly with metagenomics data. Freely avaiblable on github
|
|
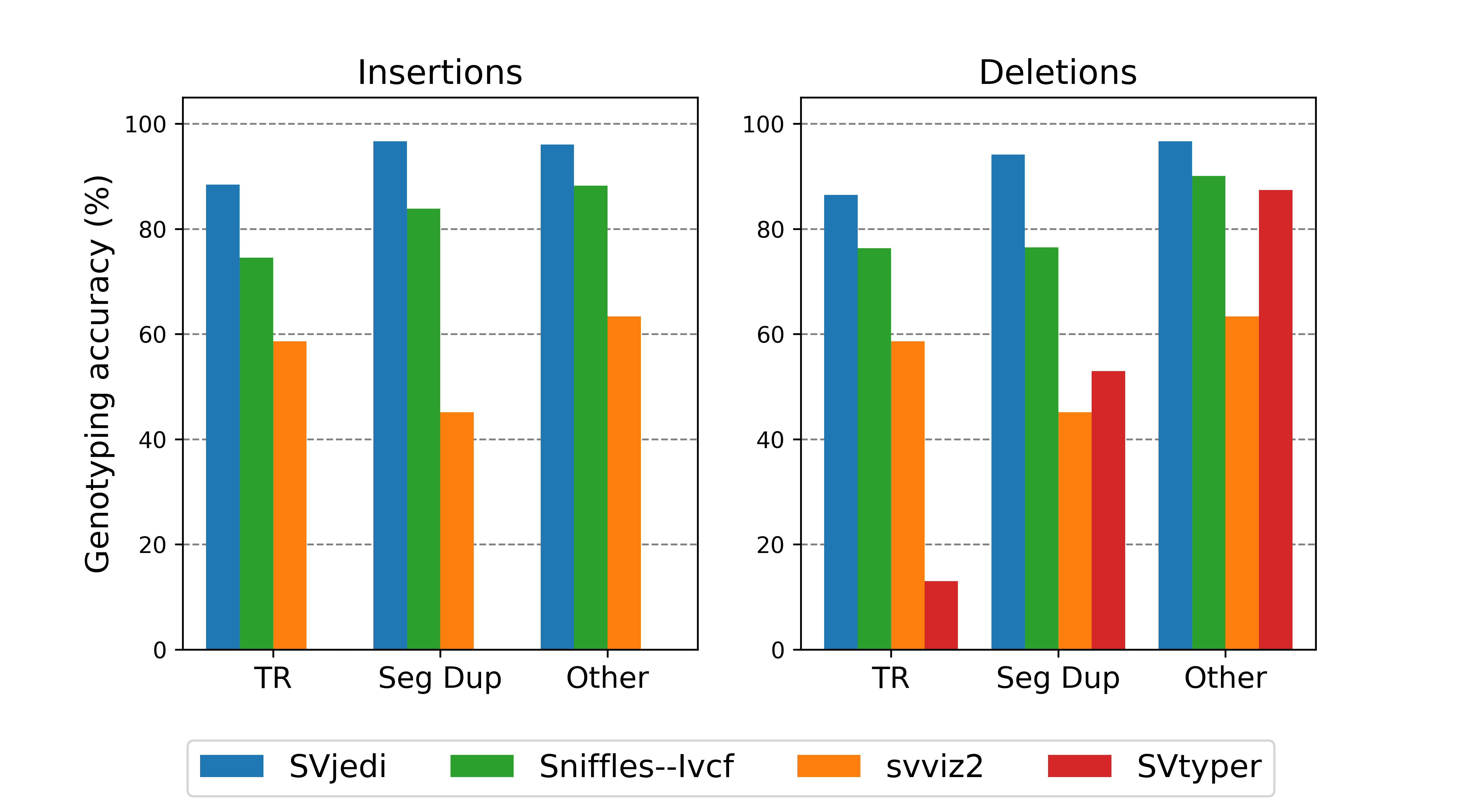
[June 2020] SVJedi finally published in Bioinformatics ! SVJedi is a new software developped by my PhD student, Lolita Lecompte, to genotype SVs with long read sequencing datasets. Freely avaiblable on github
|
|
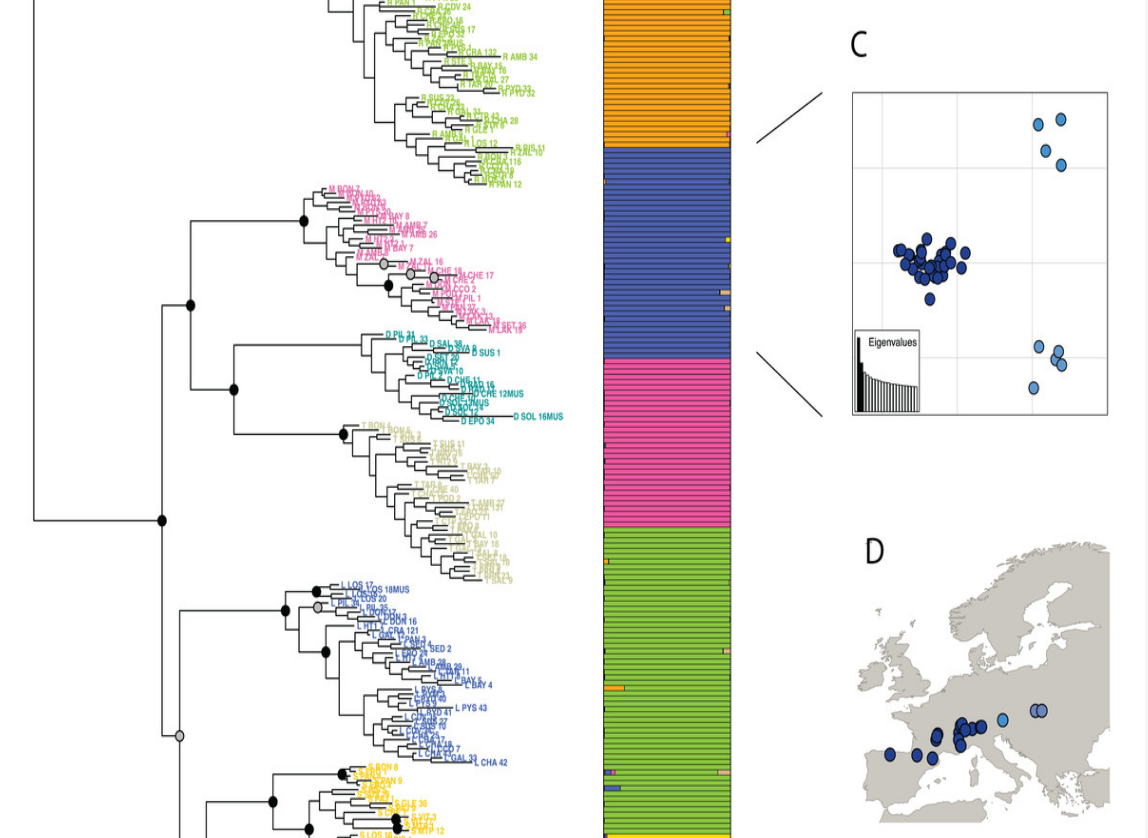
[June 2020]
DiscoSnpRad finally published in PeerJ ! DiscoSnpRAD is a software to discover SNPs and small indels in RAD-like sequencing datasets. Freely avaiblable on github
|
|
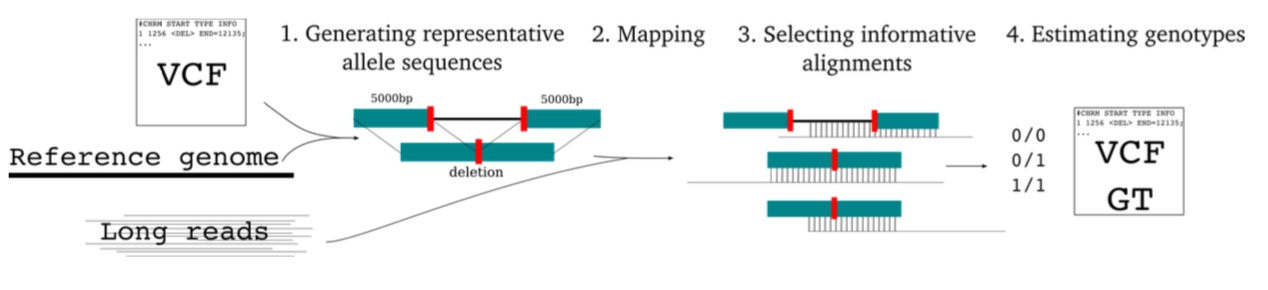
[Nov 2019]
New software : SVJedi is a new software developped by my PhD student Lolita Lecompte to genotype SVs in long read sequencing datasets. See also the preprint in BioRxiv, where SVJedi is applied on a real human dataset (HG002, GiaB SV callset)
|

















